Biological functions based on protein interplays
|
Roles and regulations of the ETS transcription factor ELF4/MEF
Mary Ann Suico, Tsuyoshi Shuto, and Hirofumi KaiJ Mol Cell Biol, Volume 9, Issue 3, June 2017, 168-177, https://doi.org/10.1093/jmcb/mjw051Abstract | Full Text |
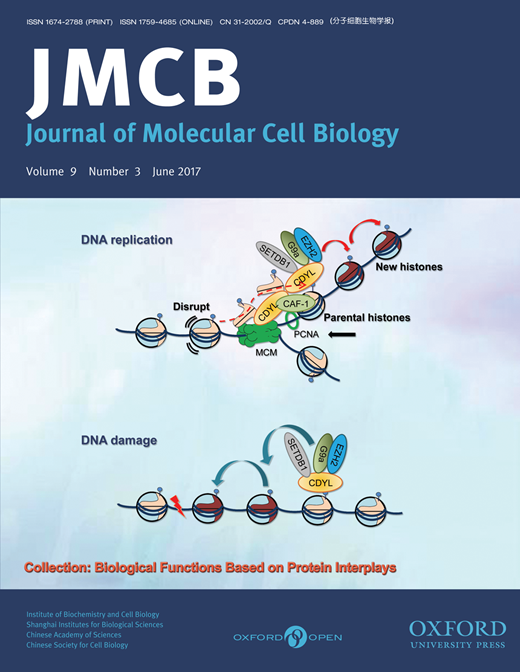
Chromodomain protein CDYL is required for transmission/restoration of repressive histone marks
|
A CK2–RNF4 interplay coordinates non-canonical SUMOylation and degradation of nuclear receptor FXR
|
BCCIPβ modulates the ribosomal and extraribosomal function of S7 through a direct interaction
Qian Ba, Xiaoguang Li, Chao Huang, Junyang Li, Yijing Fu, Peizhan Chen, Juan Duan, Miao Hao, Yinghua Zhang, Jingquan Li, Chuanqi Sun, Hao Ying, Haiyun Song, Ruiwen Zhang, Zhiyuan Shen, and Hui WangJ Mol Cell Biol, Volume 9, Issue 3, June 2017, 209-219, https://doi.org/10.1093/jmcb/mjx019Abstract | Full Text |
Human apo-SRP72 and SRP68/72 complex structures reveal the molecular basis of protein translocation
Yina Gao, Qi Zhang, Yue Lang, Yang Liu, Xiaofei Dong, Zhenhang Chen, Wenli Tian,Jun Tang, Wei Wu, Yufeng Tong, and Zhongzhou ChenJ Mol Cell Biol, Volume 9, Issue 3, June 2017, 220-230, https://doi.org/10.1093/jmcb/mjx010Abstract | Full Text |
Proteolytic cleavage is required for functional neuroligin 2 maturation and trafficking in Drosophila
Renjun Tu, Jinjun Qian, Menglong Rui, Nana Tao, Mingkuan Sun, Yan Zhuang, Huihui Lv,Junhai Han, Moyi Li, and Wei XieJ Mol Cell Biol, Volume 9, Issue 3, June 2017, 231-242, https://doi.org/10.1093/jmcb/mjx015Abstract | Full Text |
SNORD126 promotes HCC and CRC cell growth by activating the PI3K–AKT pathway through FGFR2
|
A sequence of 28S rRNA-derived small RNAs is enriched in mature sperm and various somatic tissues and possibly associates with inflammation
|