Innovative methods and techniques for frontiers of life science
|
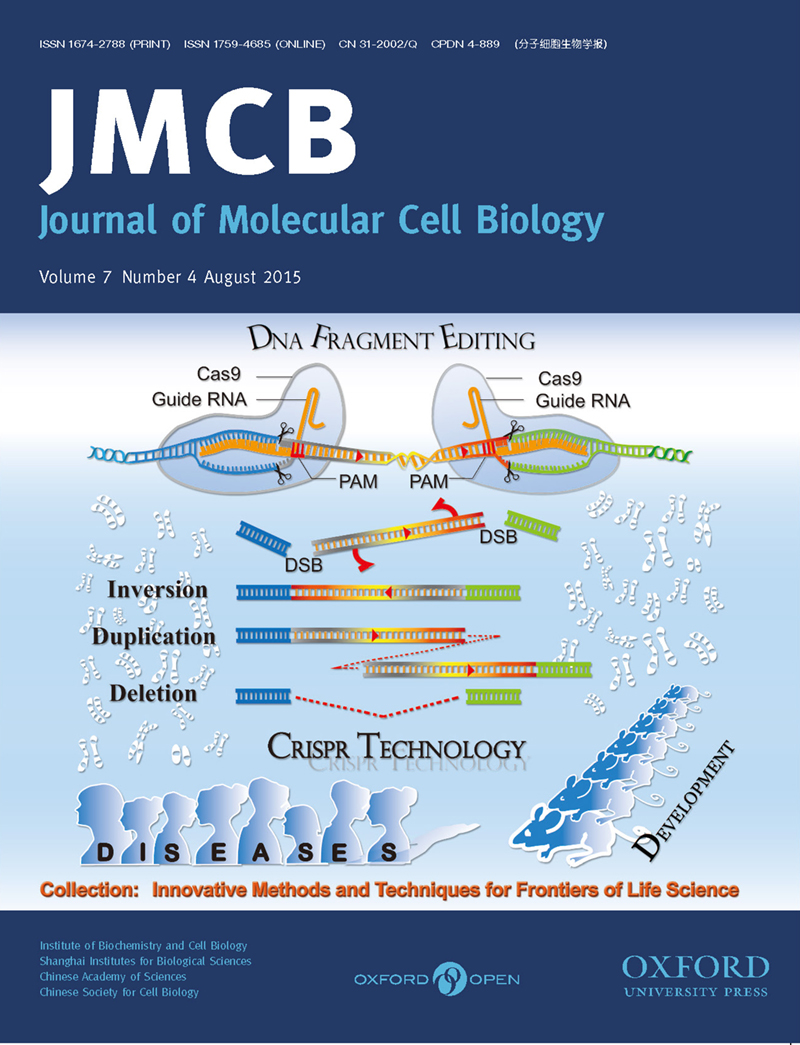
Efficient inversions and duplications of mammalian regulatory DNA elements and gene clusters by CRISPR/Cas9
Jinhuan Li, Jia Shou, Ya Guo, Yuanxiao Tang, Yonghu Wu, Zhilian Jia, Yanan Zhai, Zhifeng Chen, Quan Xu, and Qiang WuJ Mol Cell Biol, Volume 7, Issue 4, August 2015, 284-298, https://doi.org/10.1093/jmcb/mjv016Abstract | Full Text |
MBRidge: an accurate and cost-effective method for profiling DNA methylome at single-base resolution
|
Integrative analyses of RNA editing, alternative splicing, and expression of young genes in human brain transcriptome by deep RNA sequencing
|
Durable pluripotency and haploidy in epiblast stem cells derived from haploid embryonic stem cells in vitro
|
PTPROt maintains T cell immunity in the microenvironment of hepatocellular carcinoma
|
Caspase-1 activation by NLRP3 inflammasome dampens IL-33-dependent house dust mite-induced allergic lung inflammation
|
Hot spots of DNA double-strand breaks and genomic contacts of human rDNA units are involved in epigenetic regulation
Nickolai A. Tchurikov, Daria M. Fedoseeva, Dmitri V. Sosin, Anastasia V. Snezhkina, Nataliya V. Melnikova, Anna V. Kudryavtseva, Yuri V. Kravatsky, and Olga V. KretovaJ Mol Cell Biol, Volume 7, Issue 4, August 2015, 366-382, https://doi.org/10.1093/jmcb/mju038Abstract | Full Text |
Quantitative detection of in vivo physical protein–protein interactions based on DNA sequence recognition
|